Phylomapping and Comparative Genomics
Tapping into the diversity of life to inform mechanisms underlying shared traits.
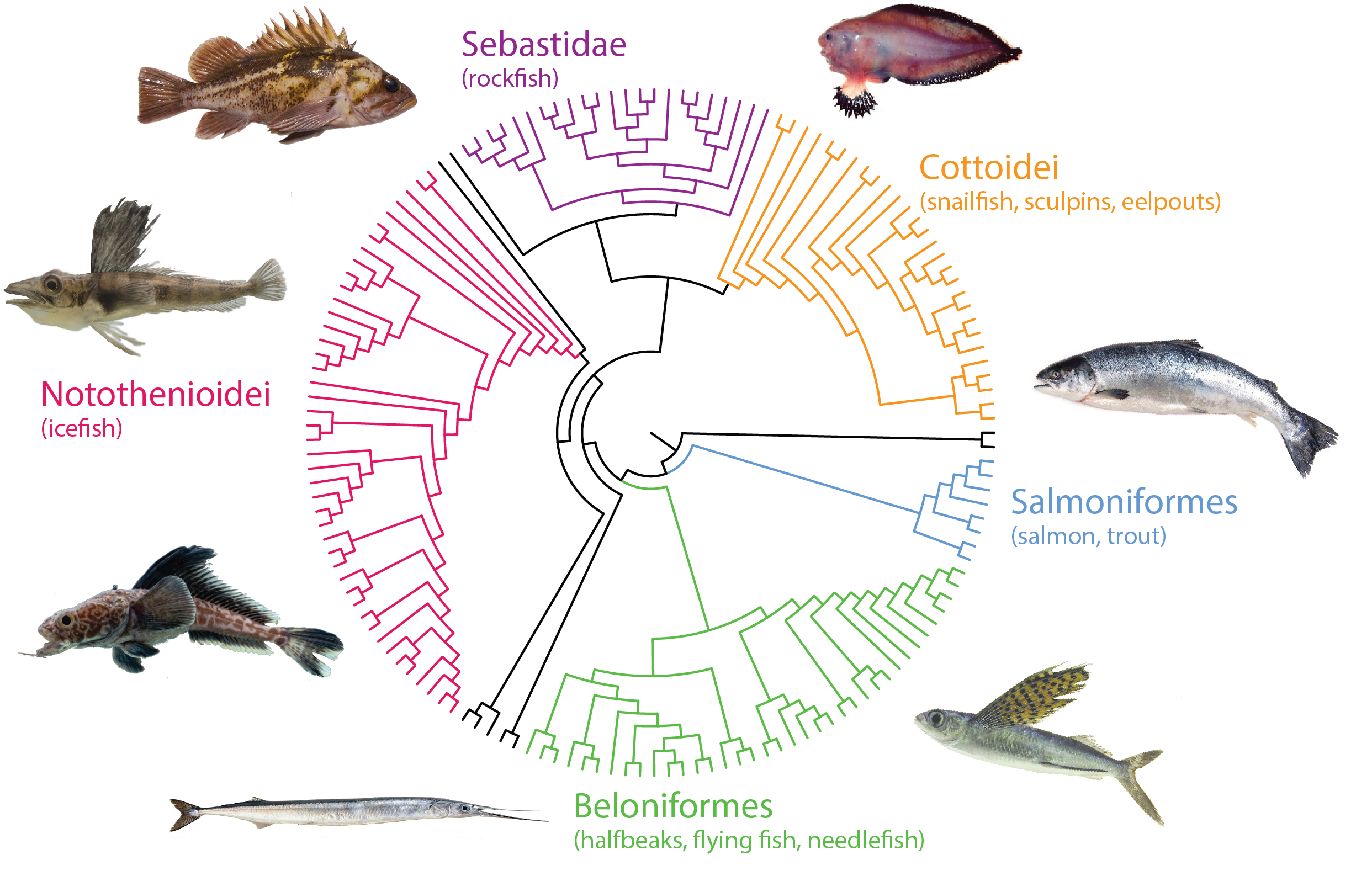
Through targeted capture approaches including over 250,000 unique baits covering both coding and conserved non coding elements, we systematically have isolated and annotated genome wide sequence complement and variation across over 250 different species of fishes. Through analysis of particular clades having fixed unique, defining, characters, we use our phylomapping approach to identify genetic signatures associating with the trait during evolution. We subsequently model these changes in zebrafish, mouse, or compare directly with variation in long-lived human populations.
Related Projects
Evolution in the extremes
Lessons in development and climate resiliency.
Comparative genomics of longevity
The tortoise and the hare and the grim reaper. How some species keep ahead, while others get caught early.
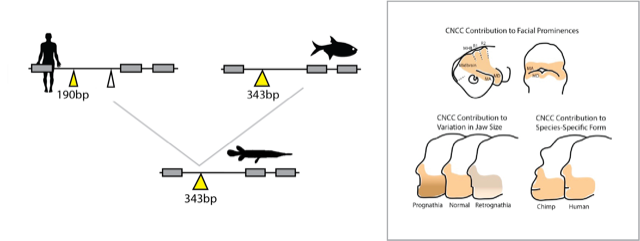
Origins and proportions of the jaw
Taking a bite out of time.
Shifting size and form
Different shapes for different capes.
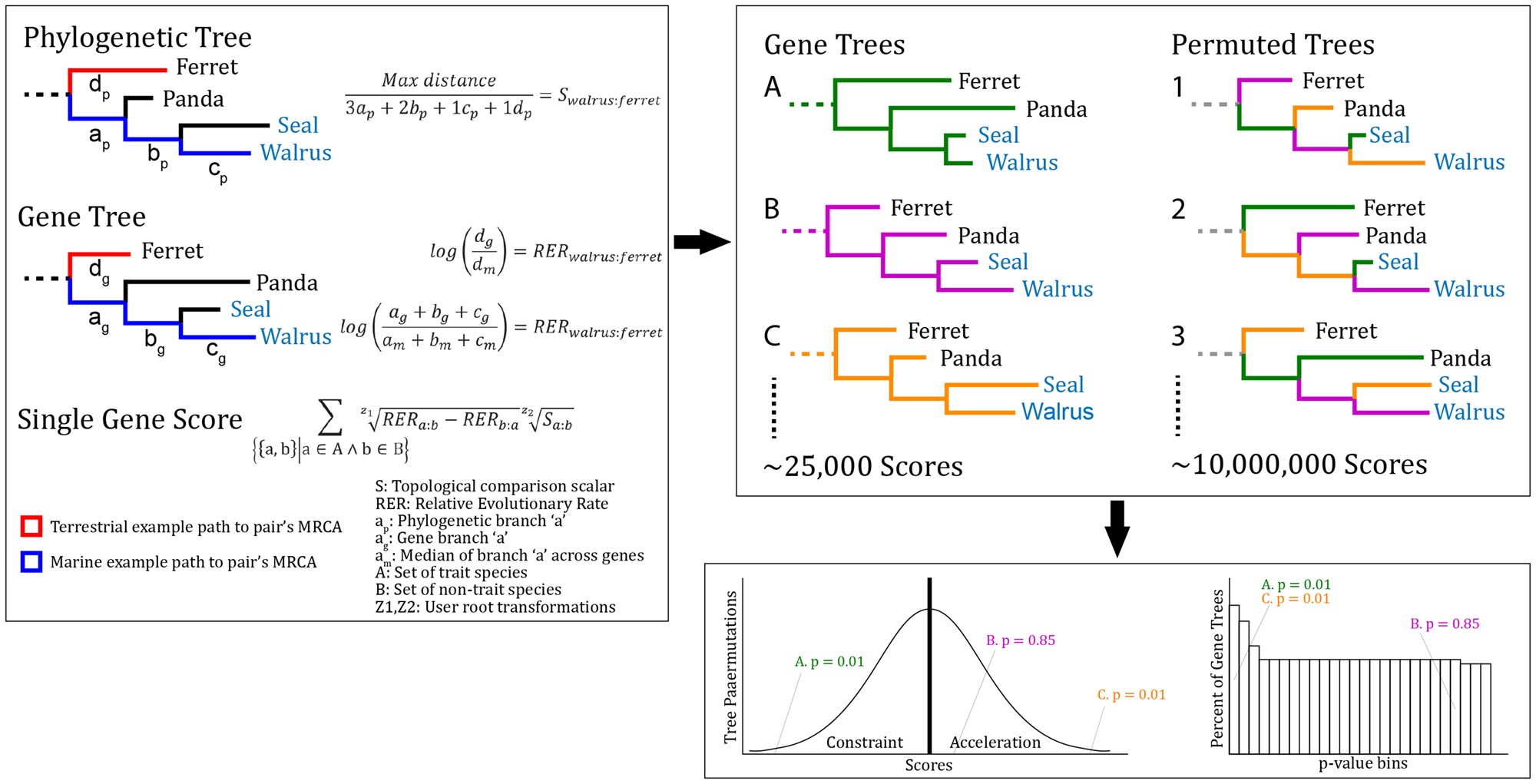
TRACCER: tool for relative evolutionary rate convergence
How to associate genes with functions through shared occurences of traits.
Key Publications
- Treaster S., Deelen J., Daane J., Murabito J., Karasik K., Harris MP. (2023) Convergent genomics of longevity in rockfishes highlights the genetics of human life span variation. Science Advances 2023 Jan 11;9(2):eadd2743[link]
- Daane, JM, Lanni J, Blum N, Iovine MK, Higdon CS, Johnson SL, Lovejoy NR, Harris MP (2021) Modulation of bioelectric cues in the evolution of flying fishes. Current Biology. Sep 13;S0960-9822(21)01190-8. doi: 10.1016/j.cub.2021.08.054
- Daane JM, Dauberg A, Hawkins MB, Smits, P., Warman M, Near T, Detrich W, Harris MP. (2019). Phylogenomic evidence of genetic changes that presage radiation of Antarctic notothenioids. Nature Eco Evol. Jul;3(7):1102-1109. PMID: 31182814
- Daane JM, Rohner N, Konstantinidis P, Djuranovic S, and Harris MP. (2016) Phylogenomic evidence for parallelism and correlative selection during fish skeletal evolution. Molecular Biology and Evolution, Jan;33(1):162-73. PMID: 26452532